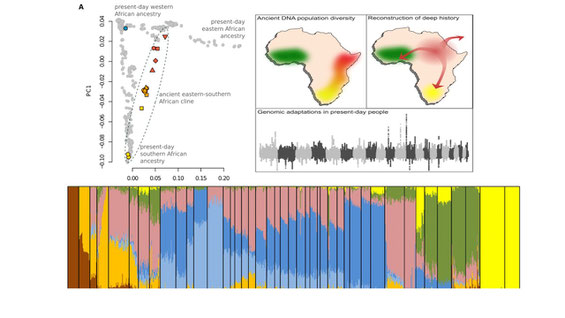
Population genomics using ancient DNA data
Dates
22nd-26th September 2025
To foster international participation, this course will be held online
overview
In recent years, novel technology for obtaining DNA molecules from archeological material has opened new horizons in the development of the field of paleogenomics. In this course, we will explore how ancient DNA (aDNA) data is generated and analyzed in population genomic research. Course content will focus on eukaryote genomes, in particular the human genome, for which a plethora of aDNA data is publicly available. On the first day of the course, participants will learn about the differences between aDNA and DNA obtained from archeological material, and how to control for DNA degradation. From Day 2 through Day 5, participants will learn about the theory and application of the main statistical methods used to analyze aDNA data. The course is structured as a combination of lectures and hands-on exercises, which will be contextualized with real research questions. Hands-on exercises will be run in a Linux environment and visualization will be run in R using RStudio.
Target audience
The course is aimed at graduate students and researchers in genetics, ecology, and anthropology who are interested in using aDNA data for population genetics inference. The course is focused on computational methods for aDNA data processing and analysis. We will go only briefly over the laboratory methods used to extract aDNA and prepare genomic libraries for sequencing. Although the examples discussed in class will focus on humans, all methods discussed in class will be relevant to those who study other organisms for which aDNA data is available.
Prerequisites
Participants should have a background in genetics, genomics, and/or population genetics, be comfortable with Unix and R, and be familiar with Next Generation Sequencing data (any organism). We ask participants without previous experience in Unix and R to go through suggested tutorials on their own before the course. No previous experience with aDNA is required.
Requirements
Attendants should have basic knowledge of UNIX and will need to use the command line on their laptops. Familiarity with a scripting language such as Python, or R will be helpful but is not
required.
program
Monday – Arun - 2-8 PM Berlin time
Data Processing and Authentication
-
Introduction to ancient DNA (aDNA) analysis
-
Processing genomic raw sequence data
-
Authentication of aDNA samples: identifying contamination & assessing postmortem DNA damage
-
Pseudo-haploid variant calling from low-coverage aDNA data
-
Hands-on exercises
Tuesday – Eduardo - 2-8 PM Berlin time
Population Structure
-
Understanding population structure
-
Principal Component Analysis (PCA) and Multidimensional Scaling (MDS)
-
Identification of genetic clusters and population structure
-
Case studies and practical exercises
Wednesday – Arun - 2-8 PM Berlin time
Introduction to the Coalescent; Demographic Inference
-
Introduction to coalescent theory
-
Estimating effective population size changes over time
-
Analyzing temporal and spatial patterns of population dynamics
-
Methods for detecting population bottlenecks and expansions
-
Reconstruction of historical population demography
-
Case studies and practical applications
Thursday – Eduardo - 2-8 PM Berlin time
D and F Statistics ; Relatedness
-
Understanding the principles and applications of D and F statistics
-
Assessing population affinity and gene flow with aDNA data
-
Analysis of relatedness and kinship with aDNA
-
Hands-on exercises and interpretation of results
Friday – Arun & Eduardo - 2-8 PM Berlin time
Simulations; Natural Selection (time permitting); Discussion
-
Introduction to population genetics simulations
-
Simulating aDNA datasets for hypothesis testing
-
Detecting signals of natural selection with aDNA data
-
Hands-on exercises, open discussion on aDNA analyses, participant projects
Cost overview
Cancellation Policy:
> 30 days before the start date = 30% cancellation fee
< 30 days before the start date= No Refund.
Physalia-courses cannot be held responsible for any travel fees, accommodation or other expenses incurred to you as a result of the cancellation.