Food Traceability Using Genomic Tools
Dates
25-28 February 2025
To foster international participation, this course will be held online
Course overview
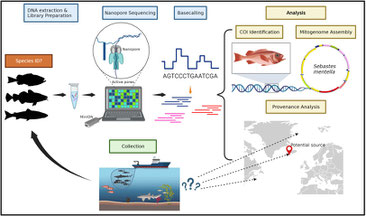
Seafood and wildlife is most traded commodities worldwide, with an estimated annual value of US$676.20 billion. Despite rigorous efforts to regulate this trade, numerous challenges continue to
emerge, complicating the enforcement and sustainable management of these critical resources.
In this intensive course, participants will receive an overview of the seafood industry, including the evolution of regulatory enforcement in the collection and sale of seafood products. The core
focus will be on introducing genomic tools used for seafood traceability. The course will introduce targeted techniques such as Sanger sequencing and quantitative PCR, then advances to the latest
in sequencing technologies (Illumina and Nanopore), bioinformatic analysis, and their practical applications in ensuring the integrity and safety of food products. By integrating theoretical
knowledge with practical workshops, this course aims to empower participants with the essential skills required for efficient food traceability.
Target audience and assumed background
Researchers and professionals in the food industry, including food safety officers, quality control technicians, and supply chain managers.
Background required: Basic understanding of molecular biology principles. Familiarity with genomic concepts and some experience with data analysis software is beneficial but not necessary.
Session content
Tuesday– Classes from 2-8 PM Berlin time – Understanding Food Traceability
Session 1: Introduction
- Introductions, expectations, and Overview of the course program
- Introduction to food traceability (ethics and consumer transparency); significance in seafood industry: the DNA revolution
In this opening session, participants will be introduced to the critical concepts of seafood traceability and authenticity. We will explore the ethical considerations and the paramount importance
of transparency for consumers. The session will highlight the role of the seafood industry in meeting global demand and discuss the evolution of DNA-based technologies aimed at protecting
consumers, producers, and aquatic resources. This comprehensive overview sets the stage for an in-depth exploration of food traceability within the seafood sector.
Session 2: Traditional approaches
- Repertoire of genetic approaches
- Exploring genetic datasets (sanger sequences, databases)
In this session, we will examine traditional genetic approaches used for identifying food products. This exploration will cover a range of basic DNA-based techniques, including Sanger sequences,
and the utilisation of genetic databases, such as Barcode of Life Data System (BOLD) and National Center for Biotechnology Information (NCBI). Participants will engage in a hands-on session
aimed at using DNA sequences to identify products of unknown identity, gaining practical experience in the process. We will also cover the basics of marker performance and introduce the databases
essential for this type of analysis. This session is designed to provide participants with a foundational understanding of how DNA technologies are applied in the traceability and authenticity of
food products, particularly within the seafood industry.
Wednesday – Classes from 2-8 PM Berlin time – Sequencing Technologies
Session 3: Experimental design
- Sampling design and laboratory protocols
- Linux basics for bioinformatics; introduction to NGS.
In this session, we will explore both traditional and next-generation sequencing (NGS) approaches, focusing on the methodologies employed and the effective application of NGS techniques,
particularly those involving Illumina platforms. Discussion will extend to best practices for sample collection and laboratory protocols, emphasising the importance of rigorous design in
experimental work. Following this, participants will have hands-on practical introduction to Linux, learning key commands essential for bioinformatics analyses covered in the course. This session
aims to equip participants with a thorough understanding of experimental design principles in the context of genetic analysis, alongside foundational skills in bioinformatics necessary for
navigating the complexities of NGS data.
Session 4: Illumina technology
- Illumina sequencing; hands-on DNA metabarcoding
- Analysis of identity
In this session, participants will learn about Illumina sequencing technology with a focus on DNA metabarcoding. Participants will learn how to manage and analyse amplicon
data generated by Illumina platforms. This includes a comprehensive walk-through of the OBITools pipeline, which covers the processing of raw Illumina reads from initial quality control through
merging, chimera detection, clustering, and taxonomic assignment. Additionally, participants will be guided on how to filter data effectively for the identification of products. By the end of
this session, participants will have gained hands-on experience and the necessary skills to handle and interpret Illumina sequencing data, equipping them with the knowledge to apply these
techniques in the field of food authenticity and traceability.
Thursday– Classes from 2-8 PM Berlin time – Advanced Sequence and Data Analysis
Session 5: Introduction to Nanopore: portability
- Food traceability and point of origin testing
- Nanopore sequencing technology; differences and applications
In this session, we will explore the cutting-edge and portable Nanopore sequencing technology, highlighting its applicability in food traceability and point of origin
testing. Our focus will shift towards developing a deeper understanding necessary for identifying species but also determining the provenance of food products. This involves an introduction to
population genetic databases and the selection of appropriate markers that are critical for tracing the origins accurately.
Following this foundational knowledge, we introduce a streamlined approach that circumvents traditional, more complex analysis methods such as PCR, qPCR, and Lateral Flow Assay (LFA), by directly
sequencing genomic DNA. This innovative session aims to provide participants with the skills to leverage portable sequencing technologies for on-site processing and immediate analysis, thereby
enhancing the efficiency and accuracy of food traceability efforts. Participants will be introduced to laboratory methods essential for generating genomic data using Nanopore technology.
Session 6: Nanopore data handling
- Bioinformatics for amplicon and whole genome sequencing; real-world data analysis
In this session, we explore Nanopore sequencing, focusing on the bioinformatics aspects for whole genome sequencing, along with real-world data analysis. Participants will
be guided through the process of handling the vast amount of data generated by Nanopore sequencing following two pathways. 1) a rapid data mining procedure to identify molecular markers to
identify species and 2) seeking to address more intricate questions, including the geographic origin of food products. Following this, the session will transition to a hands-on practical, where
participants will process Nanopore data.
Using a practical example, participants will learn how to extract information about the provenance of food products from the data. This session aims to provide a comprehensive understanding of
the end-to-end process, from data generation using Nanopore sequencing to the bioinformatics analysis necessary for determining the origin of food products. Through this hands-on approach,
participants will gain the skills and knowledge required to apply these techniques in the field of food traceability.
Friday– Classes from 2-8 PM Berlin time – Integration, Application, and Future Directions
Session 7: Recap
- Recap of key concepts and technologies discussed
- Choice of most appropriate tool
- Bioinformatic analysis considerations
- Roundtable on moving forward in food traceability, collaborative opportunities, and Q&A.
In the concluding session, we will revisit the key concepts and technologies discussed throughout the course. This session is designed to reinforce the foundational
knowledge and skills, with a special emphasis on bioinformatic analysis and the process of picking the right tool for the question at hand.
We will explore the critical factors to consider when choosing bioinformatic tools and methodologies for food traceability projects, ensuring participants understand how to apply these
technologies effectively in their work.
Following the recap, the session will transition to a roundtable discussion. This open forum provides an opportunity for participants to bring forward questions, insights, or challenges from
their personal projects or interests related to food traceability. It's a chance for collaborative exploration of potential opportunities in the field, fostering a community of practice among
attendees. The Q&A segment will allow for direct interaction and advice, ensuring participants leave with a clear path forward in their pursuit of advancing food
traceability.
COst overview
Cancellation Policy:
> 30 days before the start date = 30% cancellation fee
< 30 days before the start date= No Refund.
Physalia-courses cannot be held responsible for any travel fees, accommodation or other expenses incurred to you as a result of the cancellation.


