Gene Set Enrichment Analysis in R/Bioconductor
Dates
3-6 March 2025
To foster international participation, this course will be held online
Course overview
Gene set enrichment analysis (GSEA) is a basic method for biological data analysis. It is used to associate biological functions to a list of genes of interest,
which is to explain the results from a biology point of view. In this course, we will teach the use of popular GSEA tools, both for online-based tools and those implemented as R packages. We will
give a detailed introduction on a variety of methods of GSEA analysis, including overrepresentation analysis, univariate methods, multivariate methods, as well as extensions of GSEA analysis,
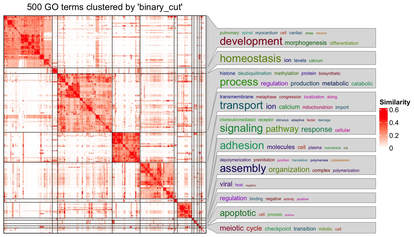
such as network-based GSEA, and single-sample GSEA. Finally, you will also learn downstream processing of GSEA results, including efficiently visualizing the massive GSEA results, clustering, and
simplifying GSEA results via various methods.
In the course, we will cover some other topics that are tightly related to GSEA analysis, such as multiple hypothesis testing. You will also learn how to implement
GSEA methods completely from scratch in R.
TARGET AUDIENCE AND ASSUMED BACKGROUND
The course is aimed at students and researchers with limited statistical knowledge and they should have a basic knowledge of R programming, e.g. basic data structures (vectors, data frames, lists).
LEARNING OUTCOMES
1. be able to perform GSEA analysis with popular tools;
2. understand various methods of GSEA analysis;
3. efficiently visualize GSEA results
Program
Sessions from 14:00 to 20:00 (Monday to Thursday, after every 50min will be a 10min break). Sessions will follow a learn-by-practice mode. After every topic will be discussion, Q&A, and practice. Students are encouraged to bring their data to the practice.
Monday– Classes from 2-8 PM Berlin time
The first day will give you an overview of what is GSEA analysis. We will teach the basic methods of GSEA analysis, the so-called overrepresentation analysis and “gene set analysis” (GSA). We will demonstrate how to use some of the popular tools which are on-line based and standalone as R packages. Thus, on the first day, you should be able to perform GSEA analysis.
Tuesday– Classes from 2-8 PM Berlin time
This day will be the day of theories. We will give a detailed introduction of the general framework of GSEA analysis, which contains univariate methods and multivariate methods. We will
demonstrate tools for the different methods. We
also learn, following the framework, how to implement a new GSEA method totally from scratch in R.
Wednesday– Classes from 2-8 PM Berlin time
We will learn extensions of the GSEA analysis, which are: 1) network-based GSEA (mainly for pathway enrichment); 2) genomic region-based GSEA; 3) GOseq; 4) single-sample GSEA. We will demonstrate the use of the corresponding tools.
Thursday– Classes from 2-8 PM Berlin time
On the last day, we will learn the downstream processing on GSEA results, which is, how to simplify the interpretation of the GSEA results. This includes efficient visualization of GSEA results, and clustering/simplifying the GSEA results.
Cost overview
Should you have any further questions, please send an email to info@physalia-courses.org
Cancellation Policy:
> 30 days before the start date = 30% cancellation fee
< 30 days before the start date= No Refund.
Physalia-courses cannot be held responsible for any travel fees, accommodation or other expenses incurred to you as a result of the cancellation.

